The Canadian agriculture industry is striving to balance both sustainability and productivity, targeting net zero emissions by 2050 and a doubling in agriculture productivity by 2030 (Government of Canada 2024). Within beef cattle production in Canada, 53-54% of greenhouse gases (GHG) are from enteric methane emissions (Basarab et al. 2012). Therefore, measurement of beef cattle methane emissions is critical for the understanding of how feed additives, vaccines, diet quality, hybrid vigour, genetic selection for feed efficiency and other best management practices for mitigating methane interact with economically important traits. We can measure these enteric gas emissions from animals in several ways. For example, the GreenFeed Emissions Monitoring (GEM) System™, takes measures of methane, carbon dioxide and hydrogen from respired and eructated (burps) gases of cattle during voluntary visitations to the system. While the GEM system provides automated and noninvasive collection of enteric gases, it is expensive specialized equipment that limits its use to primarily research sites. As such, low cost proxy measures can allow for large data collections at multiple sites. Expanding the collection of enteric methane measures will improve methodologies for reducing GHG from beef production. This could be useful when applied to development and validation of genomic breeding values and other mitigation strategies. This article will give some detail on how we are approaching this problem in our project.
The project “Reducing Greenhouse Gas Emissions from the Canadian Beef Industry through the Development and Adoption of Genomic Tools” with Livestock Gentec CEO Dr. John Basarab as PI, has been mentioned in previous articles [In the field Dec 2024; Livestock Gentec March 2025 Research Update; June 2025 NSERC pilot]. A project goal is to develop a methane emission prediction equation using Near Infrared spectroscopy (NIRS) conducted on dried and ground fecal samples.
NIRS measures the absorption of energy across the near infrared wavelengths (700-2500nm) of the electromagnetic spectrum. The absorption value from NIRS at certain wavelengths are associated with bend and stretch characteristics of various chemical bonds in a sample. We can use this variation to develop equations that predict useful attributes of the sample. For example: digestibility of of feed sample, moisture content in grain, protein content in milk, and fat content in ground meats have all shown good prediction and form the basis for industry quality testing.
In our case, we are analyzing NIRS of fecal samples to predict methane emissions. Enteric gas emissions in beef cattle are related to the fermentation of feed by rumen microbes. For example, archea microorganisms are a group of methanogens in the rumen associated with methane production. We believe we can utilize these spectra to predict GHG enteric emissions, based on the composition of metabolites in the feces. Project collaborator Dr. Amélie Vanlierde from Walloon Agricultural Research Centre in Belgium conducted similar research with fecal NIRS and observed a moderate prediction accuracy of fecal NIRS and beef and dairy cattle enteric emissions (Vanlierde et al. 2022).
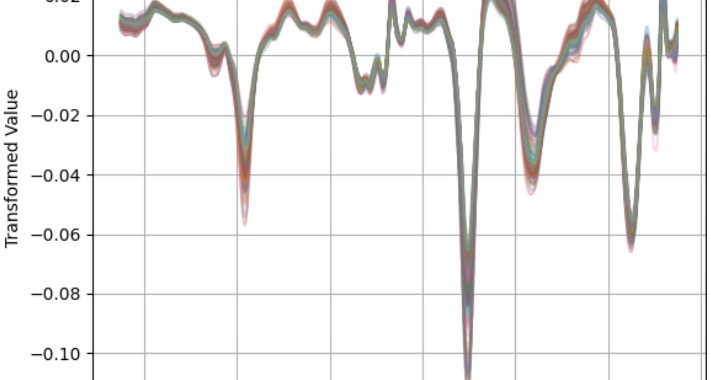
NIRS data are rich in information about sample component composition. However, the data can be noisy with interference caused by factors unrelated to composition. These factors include attributes such as particle size and moisture content. Our mitigation for this is consistent methodology across sites and fecal processing. For fecal processing, samples are dried and ground to one mm to improve the consistency of the material for spectra collection. We also conduct a series of mathematical transformations to spectra to mitigate these noise factors. Both physical and statistical processing can be applied to improve the quality of NIR spectra data for analysis.
NIRS can collect absorption values every 0.5nm wavelength. This means there can be as many as 4200 absorption values for each sample. There is rich literature exploring the best statistical methods and evaluation techniques for NIRS data analysis. Researchers commonly use Partial Least Squares Regression (PLSR) to link an attribute to NIRS spectra. We can validate results with cross-validation or a training-test set split approaches.
Figure 1: pre and post transformation spectra following SNV-D + first derivative transformation.
Finally, there are questions about the scalability of fecal NIRS as a proxy. Fecal drying and grinding, along with laboratory grade NIRS equipment which are expensive create barriers to data collection. These are good practices for the best quality data; however, there is value in exploring a more field-ready approach to expand the scope of potential data collection. To approach this goal, we are also collecting spectra using a portable NIRS on fresh fecal samples. In an ideal case, this would allow us to collect this data ‘chute-side’ at high volume.
Currently we have collected over 1350 spectra on 267 animals across four trials at three sites (Roy Berg Kinsella Research Station; Lakeland College and the Maritime Bull Test Station). Trials at Olds College Technology Access Centre for Livestock Production led by Sean Thompson are also underway. We will continue to run trials through 2028 and plan to add data from Dr. Gabriel Ribeiro at the University of Saskatchewan. These trials will include cows, heifers, bulls, native steers, and beef-on-dairy steers. Research sites will use a variety of feeds including high forage-based rations and high-grain finishing diets. This will help to produce a large dataset with high variability. The large and diverse dataset will help us to evaluate these NIRS based enteric emission predictions.
Figure 2: Labelled fecal samples collected from Lakeland College by Dr. Obioha Duranna and team.

Citations:
Basarab, J., V. Baron, O. López-Campos, J. Aalhus, K. Haugen-Kozyra, E. Okine. 2012.
Greenhouse gas emissions from calf- and yearling-fed beef production systems, with and without the use of growth promotants. Animals, 2:195–220.
Government of Canada 2024. Sustainable Development Goal 2: Zero hunger. https://www.canada.ca/en/employment-social-development/programs/agenda-2030/zero-hunger.html [accessed August 27, 2025].
Vanlierde, A, F. Dehareng, A. Mertens, M. Mathot, A. Lefevre, et al 2022. Estimation of methane eructed by dairy and beef cattle using faecal near-infrared spectra. 73. Annual Meeting of the European Federation of Animal Science, EAAP, Sep 2022, Porto, Portugal. pp.333. ⟨hal-04792528⟩